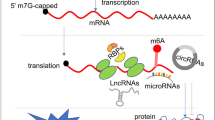
The regulation of gene expression is fundamental for life. Whereas the role of transcriptional regulation of gene expression has been studied for several decades, it has been clear over the past two decades that post-transcriptional regulation of gene expression, of which translation regulation is a major part, can be equally important. Translation can be divided into four main stages: initiation, elongation, termination and ribosome recycling. Translation is controlled mainly during its initiation, a process which culminates in a ribosome positioned with an initiator tRNA over the start codon and, thus, ready to begin elongation of the protein chain. mRNA translation has emerged as a powerful tool for the development of innovative therapies, yet the detailed mechanisms underlying the complex process of initiation remain unclear. Recent studies in yeast and mammals have started to shed light on some previously unclear aspects of this process. In this Review, we discuss the current state of knowledge on eukaryotic translation initiation and its regulation in health and disease. Specifically, we focus on recent advances in understanding the processes involved in assembling the 43S pre-initiation complex and its recruitment by the cap-binding complex eukaryotic translation initiation factor 4F (eIF4F) at the 5′ end of mRNA. In addition, we discuss recent insights into ribosome scanning along the 5′ untranslated region of mRNA and selection of the start codon, which culminates in joining of the 60S large subunit and formation of the 80S initiation complex.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
206,07 € per year
only 17,17 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others

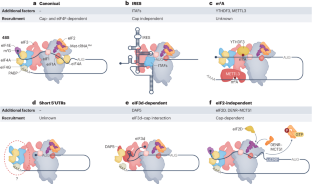
eIF4E-independent translation is largely eIF3d-dependent
Article Open access 06 August 2024

eIF5B gates the transition from translation initiation to elongation
Article 18 September 2019
The regulation of protein translation and its implications for cancer
Article Open access 18 February 2021
References
- Harnett, D. et al. A critical period of translational control during brain development at codon resolution. Nat. Struct. Mol. Biol.29, 1277–1290 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature473, 337–342 (2011). ArticleADSPubMedGoogle Scholar
- Byrne, R., Levin, J. G., Bladen, H. A. & Nirenberg, M. W. The in vitro formation of a DNA–ribosome complex. Proc. Natl Acad. Sci. USA52, 140–148 (1964). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Kohler, R., Mooney, R. A., Mills, D. J., Landick, R. & Cramer, P. Architecture of a transcribing–translating expressome. Science356, 194–197 (2017). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Wang, C. et al. Structural basis of transcription–translation coupling. Science369, 1359–1365 (2020). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Webster, M. W. et al. Structural basis of transcription–translation coupling and collision in bacteria. Science369, 1355–1359 (2020). ArticleADSCASPubMedGoogle Scholar
- Aitken, C. E. & Lorsch, J. R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol.19, 568–576 (2012). ArticleCASPubMedGoogle Scholar
- Hashem, Y. & Frank, J. The jigsaw puzzle of mRNA translation initiation in eukaryotes: a decade of structures unraveling the mechanics of the process. Annu. Rev. Biophys.https://doi.org/10.1146/annurev-biophys-070816-034034 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Hinnebusch, A. G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem.83, 779–812 (2014). ArticleCASPubMedGoogle Scholar
- Valášek, L. S. et al. Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res.45, 10948–10968 (2017). ArticlePubMedPubMed CentralGoogle Scholar
- Bohlen, J., Fenzl, K., Kramer, G., Bukau, B. & Teleman, A. A. Selective 40S footprinting reveals cap-tethered ribosome scanning in human cells. Mol. Cellhttps://doi.org/10.1016/j.molcel.2020.06.005 (2020). This study presents the first clear evidence that scanning can be cap-tethered in human cells.ArticlePubMedGoogle Scholar
- Brito Querido, J. et al. Structure of a human 48S translational initiation complex. Science369, 1220–1227 (2020). This structure reveals how eIF4F interacts with the 43S complex.ArticleADSCASPubMedGoogle Scholar
- Chiluiza, D., Bargo, S., Callahan, R. & Rhoads, R. E. Expression of truncated eukaryotic initiation factor 3e (eIF3e) resulting from integration of mouse mammary tumor virus (MMTV) causes a shift from cap-dependent to cap-independent translation. J. Biol. Chem.286, 31288–31296 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Grifo, J. A., Tahara, S. M., Morgan, M. A., Shatkin, A. J. & Merrick, W. C. New initiation factor activity required for globin mRNA translation. J. Biol. Chem.258, 5804–5810 (1983). ArticleCASPubMedGoogle Scholar
- Kumar, P., Hellen, C. U. T. & Pestova, T. V. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev.30, 1573–1588 (2016). This study presents strong evidence to support the threading model of mRNA recruitment to the 43S complex in mammals.ArticleCASPubMedPubMed CentralGoogle Scholar
- Llácer, J. L. et al. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell59, 399–412 (2015). ArticlePubMedPubMed CentralGoogle Scholar
- Marintchev, A. et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell136, 447–460 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pestova, T. V. & Kolupaeva, V. G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev.16, 2906–2922 (2002). ArticleCASPubMedPubMed CentralGoogle Scholar
- Brito Querido, J. et al. The structure of a human translation initiation complex reveals two independent roles for the helicase eIF4A. Nat. Struct. Mol. Biol.https://doi.org/10.1038/s41594-023-01196-0 (2024). This study reveals that in addition to the eIF4A molecule that is part of the eIF4F complex, there is a second molecule of eIF4A in the 48S complex, which functions separately from eIF4F.
- Villa, N., Do, A., Hershey, J. W. B. & Fraser, C. S. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem.288, 32932–32940 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Berthelot, K., Muldoon, M., Rajkowitsch, L., Hughes, J. & McCarthy, J. E. G. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol.51, 987–1001 (2004). ArticleCASPubMedGoogle Scholar
- Kozak, M. Role of ATP in binding and migration of 40S ribosomal subunits. Cell22, 459–467 (1980). ArticleCASPubMedGoogle Scholar
- Nielsen, K. H. et al. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J.23, 1166–1177 (2004). ArticleCASPubMedPubMed CentralGoogle Scholar
- Shirokikh, N. E., Dutikova, Y. S., Staroverova, M. A., Hannan, R. D. & Preiss, T. Migration of small ribosomal subunits on the 5’ untranslated regions of capped messenger RNA. Int. J. Mol. Sci.20, 4464 (2019). ArticleCASPubMedPubMed CentralGoogle Scholar
- Simonetti, A., Guca, E., Bochler, A., Kuhn, L. & Hashem, Y. Structural insights into the mammalian late-stage initiation complexes. Cell Rep.31, 107497 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Wang, J. et al. Rapid 40S scanning and its regulation by mRNA structure during eukaryotic translation initiation. Cell185, 4474–4487 (2022). This study reveals the kinetics of scanning and finds that multiple copies of eIF4A can have a role during mRNA recruitment.ArticleCASPubMedPubMed CentralGoogle Scholar
- Yi, S.-H. et al. Conformational rearrangements upon start codon recognition in human 48S translation initiation complex. Nucleic Acids Res.50, 5282–5298 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- García-García, C., Frieda, K. L., Feoktistova, K., Fraser, C. S. & Block, S. M. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science348, 1486–1488 (2015). ArticleADSPubMedPubMed CentralGoogle Scholar
- Sen, N. D., Zhou, F., Harris, M. S., Ingolia, N. T. & Hinnebusch, A. G. eIF4B stimulates translation of long mRNAs with structured 5′ UTRs and low closed-loop potential but weak dependence on eIF4G. Proc. Natl Acad. Sci. USA113, 10464–10472 (2016). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Shahbazian, D. et al. Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol. Cell Biol.30, 1478–1485 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Calviello, L. et al. DDX3 depletion represses translation of mRNAs with complex 5′ UTRs. Nucleic Acids Res.49, 5336–5350 (2021). ArticleCASPubMedPubMed CentralGoogle Scholar
- Gupta, N., Lorsch, J. R. & Hinnebusch, A. G. Yeast Ded1 promotes 48S translation pre-initiation complex assembly in an mRNA-specific and eIF4F-dependent manner. eLife7, e38892 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Pisareva, V. P., Pisarev, A. V., Komar, A. A., Hellen, C. U. T. & Pestova, T. V. Translation initiation on mammalian mRNAs with structured 5′ UTRs requires DExH-box protein DHX29. Cell135, 1237–1250 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Huang, B. Y. & Fernández, I. S. Long-range interdomain communications in eIF5B regulate GTP hydrolysis and translation initiation. Proc. Natl Acad. Sci. USA117, 1429–1437 (2020). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Lapointe, C. P. et al. eIF5B and eIF1A reorient initiator tRNA to allow ribosomal subunit joining. Nature607, 185–190 (2022). This study reveals how eIF5B coordinates with eIF1A to reorient the tRNA and allow joining of the 60S ribosome subunit.ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Pestova, T. V. et al. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature403, 332–335 (2000). This study reveals that joining of the 60S ribosome subunit requires eIF5B.ArticleADSCASPubMedGoogle Scholar
- Wang, J. et al. eIF5B gates the transition from translation initiation to elongation. Nature573, 605–608 (2019). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Wang, J. et al. Structural basis for the transition from translation initiation to elongation by an 80S–eIF5B complex. Nat. Commun.11, 5003 (2020). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Archer, S. K., Shirokikh, N. E., Beilharz, T. H. & Preiss, T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature535, 570–574 (2016). ArticleADSCASPubMedGoogle Scholar
- Choe, J. et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature561, 556–560 (2018). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Díaz-López, I., Toribio, R., Berlanga, J. J. & Ventoso, I. An mRNA-binding channel in the ES6S region of the translation 48S-PIC promotes RNA unwinding and scanning. eLife8, e48246 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Gu, Y., Mao, Y., Jia, L., Dong, L. & Qian, S.-B. Bi-directional ribosome scanning controls the stringency of start codon selection. Nat. Commun.12, 6604 (2021). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Hayek, H. et al. eIF3 interacts with histone H4 messenger RNA to regulate its translation. J. Biol. Chem.296, 100578 (2021). ArticleCASPubMedPubMed CentralGoogle Scholar
- Herrmannová, A. et al. Adapted formaldehyde gradient cross-linking protocol implicates human eIF3d and eIF3c, k and l subunits in the 43S and 48S pre-initiation complex assembly, respectively. Nucleic Acids Res.48, 1969–1984 (2020). ArticlePubMedGoogle Scholar
- Lee, A. S., Kranzusch, P. J., Doudna, J. A. & Cate, J. H. D. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature536, 96–99 (2016). This study reveals that eIF3d is a cap-binding protein required for the translation of specific mRNAs.ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Robert, F., Cencic, R., Cai, R., Schmeing, T. M. & Pelletier, J. RNA-tethering assay and eIF4G:eIF4A obligate dimer design uncovers multiple eIF4F functional complexes. Nucleic Acids Res.48, 8562–8575 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Sokabe, M. & Fraser, C. S. A helicase-independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc. Natl Acad. Sci. USA114, 6304–6309 (2017). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Yourik, P. et al. Yeast eIF4A enhances recruitment of mRNAs regardless of their structural complexity. eLife6, e31476 (2017). ArticlePubMedPubMed CentralGoogle Scholar
- Zinshteyn, B., Rojas-Duran, M. F. & Gilbert, W. V. Translation initiation factor eIF4G1 preferentially binds yeast transcript leaders containing conserved oligo-uridine motifs. RNA23, 1365–1375 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Malik, I., Kelley, C. P., Wang, E. T. & Todd, P. K. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol.https://doi.org/10.1038/s41580-021-00382-6 (2021). ArticlePubMedPubMed CentralGoogle Scholar
- Meyer, K. D. et al. 5′ UTR m 6 A promotes cap-independent translation. Cell163, 999–1010 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhou, J. et al. Dynamic m 6 A mRNA methylation directs translational control of heat shock response. Nature526, 591–594 (2015). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Legnini, I. et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell66, 22–37.e9 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pamudurti, N. R. et al. Translation of circRNAs. Mol. Cell66, 9–21.e7 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Yang, Y. et al. Extensive translation of circular RNAs driven by N 6 -methyladenosine. Cell Res.27, 626–641 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
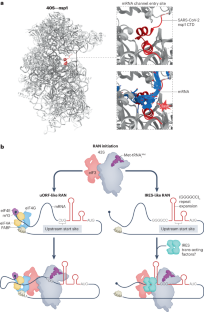
- Banerjee, A. K. et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell183, 1325–1339.e21 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Schubert, K. et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol.27, 959–966 (2020). ArticleCASPubMedGoogle Scholar
- Thoms, M. et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science369, 1249–1255 (2020). This study reveals how nsp1 of SARS-CoV-2 binds to the 40S small ribosomal subunit.ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Tidu, A. et al. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNAhttps://doi.org/10.1261/rna.078121.120 (2020). ArticlePubMedGoogle Scholar
- Chang, J. H. et al. Crystal structure of the eIF4A–PDCD4 complex. Proc. Natl Acad. Sci. USA106, 3148–3153 (2009). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Loh, P. G. et al. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J.28, 274–285 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Suzuki, C. et al. PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc. Natl Acad. Sci. USA105, 3274–3279 (2008). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Yang, H.-S. et al. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol. Cell. Biol.24, 3894–3906 (2004). ArticleCASPubMedPubMed CentralGoogle Scholar
- Passmore, L. A. et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell26, 41–50 (2007). ArticleCASPubMedGoogle Scholar
- Sokabe, M. & Fraser, C. S. Human eukaryotic initiation factor 2 (eIF2)–GTP–Met-tRNAi ternary complex and eIF3 stabilize the 43S preinitiation complex. J. Biol. Chem.289, 31827–31836 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Bochler, A. et al. Structural differences in translation initiation between pathogenic trypanosomatids and their mammalian hosts. Cell Rep.33, 108534 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Elantak, L. et al. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J. Mol. Biol.396, 1097–1116 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Fekete, C. A. et al. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J.26, 1602–1614 (2007). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hussain, T. et al. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell159, 597–607 (2014). This study identifies some crucial structural rearrangement of the 48S complex upon start-codon selection.ArticleCASPubMedPubMed CentralGoogle Scholar
- Lomakin, I. B. & Steitz, T. A. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature500, 307–311 (2013). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Nanda, J. S., Saini, A. K., Muñoz, A. M., Hinnebusch, A. G. & Lorsch, J. R. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J. Biol. Chem.288, 5316–5329 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Nanda, J. S. et al. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol.394, 268–285 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pestova, T. V., Borukhov, S. I. & Hellen, C. U. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature394, 854–859 (1998). ArticleADSCASPubMedGoogle Scholar
- Thakur, A., Marler, L. & Hinnebusch, A. G. A network of eIF2β interactions with eIF1 and Met-tRNAi promotes accurate start codon selection by the translation preinitiation complex. Nucleic Acids Res.5, 2574–2593 (2019). ArticleGoogle Scholar
- Thakur, A. & Hinnebusch, A. G. eIF1 loop 2 interactions with Met-tRNAi control the accuracy of start codon selection by the scanning preinitiation complex. Proc. Natl Acad. Sci. USA115, E4159–E4168 (2018). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Valášek, L., Nielsen, K. H., Zhang, F., Fekete, C. A. & Hinnebusch, A. G. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell Biol.24, 9437–9455 (2004). ArticlePubMedPubMed CentralGoogle Scholar
- Weisser, M., Voigts-Hoffmann, F., Rabl, J., Leibundgut, M. & Ban, N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat. Struct. Mol. Biol.20, 1015–1017 (2013). ArticleCASPubMedGoogle Scholar
- Zhou, F., Zhang, H., Kulkarni, S. D., Lorsch, J. R. & Hinnebusch, A. G. eIF1 discriminates against suboptimal initiation sites to prevent excessive uORF translation genome-wide. RNA26, 419–438 (2020). ArticlePubMedPubMed CentralGoogle Scholar
- des Georges, A. et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature525, 491–495 (2015). This study reveals the first complete architecture of mammalian eIF3.ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Erzberger, J. P. et al. Molecular architecture of the 40S ⋅ eIF1 ⋅ eIF3 translation initiation complex. Cell158, 1123–1135 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kratzat, H. et al. A structural inventory of native ribosomal ABCE1–43S pre-initiation complexes. EMBO J.40, e105179 (2020). ArticlePubMedPubMed CentralGoogle Scholar
- Llácer, J. L. et al. Large-scale movement of eIF3 domains during translation initiation modulate start codon selection. Nucleic Acids Res.49, 11491–11511 (2021). ArticlePubMedPubMed CentralGoogle Scholar
- Llácer, J. L. et al. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. eLife7, e39273 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Aitken, C. E. et al. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. eLife5, e20934 (2016). ArticlePubMedPubMed CentralGoogle Scholar
- Sun, C. et al. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res.41, 7512–7521 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Cuchalová, L. et al. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol.30, 4671–4686 (2010). ArticlePubMedPubMed CentralGoogle Scholar
- Obayashi, E. et al. Molecular landscape of the ribosome pre-initiation complex during mRNA scanning: structural role for eIF3c and its control by eIF5. Cell Rep.18, 2651–2663 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lamper, A. M., Fleming, R. H., Ladd, K. M. & Lee, A. S. Y. A phosphorylation-regulated eIF3d translation switch mediates cellular adaptation to metabolic stress. Science370, 853–856 (2020). ArticleADSCASPubMedGoogle Scholar
- Pelletier, J. & Sonenberg, N. The organizing principles of eukaryotic ribosome recruitment. Annu. Rev. Biochem.88, 307–335 (2019). ArticleCASPubMedGoogle Scholar
- Gouridis, G. et al. ABCE1 controls ribosome recycling by an asymmetric dynamic conformational equilibrium. Cell Rep.28, 723–734.e6 (2019). ArticleCASPubMedPubMed CentralGoogle Scholar
- Young, D. J. & Guydosh, N. R. Hcr1/eIF3j is a 60S ribosomal subunit recycling accessory factor in vivo. Cell Rep.28, 39–50.e4 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Aylett, C. H. S., Boehringer, D., Erzberger, J. P., Schaefer, T. & Ban, N. Structure of a yeast 40S–eIF1–eIF1A–eIF3–eIF3j initiation complex. Nat. Struct. Mol. Biol.22, 269–271 (2015). ArticleCASPubMedGoogle Scholar
- Fraser, C. S., Berry, K. E., Hershey, J. W. B. & Doudna, J. A. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol. Cell26, 811–819 (2007). ArticleCASPubMedGoogle Scholar
- Park, E.-H. et al. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1•PABP mRNPs in vivo. EMBO J.30, 302–316 (2011). ArticleCASPubMedGoogle Scholar
- Yanagiya, A. et al. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol. Cell Biol.29, 1661–1669 (2009). ArticleCASPubMedGoogle Scholar
- Haimov, O. et al. Dynamic interaction of eukaryotic initiation factor 4G1 (eIF4G1) with eIF4E and eIF1 underlies scanning-dependent and -independent translation. Mol. Cell Biol.38, e00139-18 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Kahvejian, A., Svitkin, Y. V., Sukarieh, R., M’Boutchou, M.-N. & Sonenberg, N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev.19, 104–113 (2005). ArticleCASPubMedPubMed CentralGoogle Scholar
- Amrani, N., Ghosh, S., Mangus, D. A. & Jacobson, A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature453, 1276–1280 (2008). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Lai, W.-J. C. et al. Intrinsically unstructured sequences in the mRNA 3′ UTR reduce the ability of poly(A) tail to enhance translation. J. Mol. Biol.434, 167877 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- Wells, S. E., Hillner, P. E., Vale, R. D. & Sachs, A. B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell2, 135–140 (1998). ArticleCASPubMedGoogle Scholar
- Alekhina, O. M., Terenin, I. M., Dmitriev, S. E. & Vassilenko, K. S. Functional cyclization of eukaryotic mRNAs. Int. J. Mol. Sci.21, 1677 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Vicens, Q., Kieft, J. S. & Rissland, O. S. Revisiting the closed-loop model and the nature of mRNA 5′–3′ communication. Mol. Cell72, 805–812 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Yamanaka, S. et al. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J.19, 5533–5541 (2000). ArticleCASPubMedPubMed CentralGoogle Scholar
- Liberman, N. et al. DAP5 associates with eIF2β and eIF4AI to promote internal ribosome entry site driven translation. Nucleic Acids Res.43, 3764–3775 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kulak, N. A., Pichler, G., Paron, I., Nagaraj, N. & Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods11, 319–324 (2014). ArticleCASPubMedGoogle Scholar
- Tauber, D. et al. Modulation of RNA condensation by the DEAD-Box protein eIF4A. Cell180, 411–426.e16 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Çetin, B. & O’Leary, S. E. mRNA- and factor-driven dynamic variability controls eIF4F-cap recognition for translation initiation. Nucleic Acids Res.50, 8240–8261 (2022). ArticlePubMedPubMed CentralGoogle Scholar
- Feoktistova, K., Tuvshintogs, E., Do, A. & Fraser, C. S. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc. Natl Acad. Sci. USA110, 13339–13344 (2013). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Rozovsky, N., Butterworth, A. C. & Moore, M. J. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA14, 2136–2148 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Andreou, A. Z. & Klostermeier, D. The DEAD-box helicase eIF4A. RNA Biol.10, 19–32 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Ma, X. M. & Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol.10, 307–318 (2009). ArticlePubMedGoogle Scholar
- Marcotrigiano, J., Gingras, A. C., Sonenberg, N. & Burley, S. K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell3, 707–716 (1999). ArticleCASPubMedGoogle Scholar
- Bartish, M. et al. The role of eIF4F-driven mRNA translation in regulating the tumour microenvironment. Nat. Rev. Cancerhttps://doi.org/10.1038/s41568-023-00567-5 (2023). ArticlePubMedGoogle Scholar
- Bhat, M. et al. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov.14, 261–278 (2015). ArticleCASPubMedGoogle Scholar
- Harris, T. E. et al. mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J.25, 1659–1668 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- LeFebvre, A. K. et al. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem.281, 22917–22932 (2006). ArticleCASPubMedGoogle Scholar
- Giess, A. et al. Profiling of small ribosomal subunits reveals modes and regulation of translation initiation. Cell Rep.31, 107534 (2020). ArticleCASPubMedGoogle Scholar
- Cheung, Y.-N. et al. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev.21, 1217–1230 (2007). ArticleCASPubMedPubMed CentralGoogle Scholar
- Maag, D., Fekete, C. A., Gryczynski, Z. & Lorsch, J. R. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell17, 265–275 (2005). ArticleCASPubMedGoogle Scholar
- Martin-Marcos, P. et al. Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy. RNA20, 150–167 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol.41, 262–272 (2023). ArticleCASPubMedGoogle Scholar
- Wen, S., Qadir, J. & Yang, B. B. Circular RNA translation: novel protein isoforms and clinical significance. Trends Mol. Med.https://doi.org/10.1016/j.molmed.2022.03.003 (2022). ArticlePubMedGoogle Scholar
- Wesselhoeft, R. A., Kowalski, P. S. & Anderson, D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun.9, 2629 (2018). ArticleADSPubMedPubMed CentralGoogle Scholar
- Chen, C. Y. & Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science268, 415–417 (1995). ArticleADSCASPubMedGoogle Scholar
- Mailliot, J. & Martin, F. Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdiscip. Rev. RNAhttps://doi.org/10.1002/wrna.1458 (2018). ArticlePubMedGoogle Scholar
- Gallie, D. R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev.5, 2108–2116 (1991). ArticleCASPubMedGoogle Scholar
- Leppek, K., Das, R. & Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol.19, 158–174 (2018). ArticleCASPubMedGoogle Scholar
- Elfakess, R. et al. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res.39, 7598–7609 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Elfakess, R. & Dikstein, R. A translation initiation element specific to mRNAs with very short 5′ UTR that also regulates transcription. PLoS ONE3, e3094 (2008). ArticleADSPubMedPubMed CentralGoogle Scholar
- Akulich, K. A. et al. Four translation initiation pathways employed by the leaderless mRNA in eukaryotes. Sci. Rep.6, 37905 (2016). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Andreev, D. E., Terenin, I. M., Dunaevsky, Y. E., Dmitriev, S. E. & Shatsky, I. N. A leaderless mRNA can bind to mammalian 80S ribosomes and direct polypeptide synthesis in the absence of translation initiation factors. Mol. Cell Biol.26, 3164–3169 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Cattie, D. J. et al. Mutations in nonessential eIF3k and eIF3l genes confer lifespan extension and enhanced resistance to ER stress in Caenorhabditis elegans. PLoS Genet.12, e1006326 (2016). ArticlePubMedPubMed CentralGoogle Scholar
- Smith, M. D. et al. Human-like eukaryotic translation initiation factor 3 from Neurospora crassa. PLoS ONE8, e78715 (2013). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Chapat, C. et al. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc. Natl Acad. Sci. USA114, 5425–5430 (2017). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Niederer, R. O., Rojas-Duran, M. F., Zinshteyn, B. & Gilbert, W. V. Direct analysis of ribosome targeting illuminates thousand-fold regulation of translation initiation. Cell Syst.13, 256–264.e3 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- Asano, K. et al. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J.20, 2326–2337 (2001). ArticleCASPubMedPubMed CentralGoogle Scholar
- He, H. et al. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol. Cell Biol.23, 5431–5445 (2003). ArticleCASPubMedPubMed CentralGoogle Scholar
- Luna, R. E. et al. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Rep.1, 689–702 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Walker, S. E. et al. Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA19, 191–207 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Eliseev, B. et al. Structure of a human cap-dependent 48S translation pre-initiation complex. Nucleic Acids Res.46, 2678–2689 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Padrón, A., Iwasaki, S. & Ingolia, N. T. Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell75, 875–887.e5 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Li, K., Kong, J., Zhang, S., Zhao, T. & Qian, W. Distance-dependent inhibition of translation initiation by downstream out-of-frame AUGs is consistent with a Brownian ratchet process of ribosome scanning. Genome Biol.23, 254 (2022). ArticlePubMedPubMed CentralGoogle Scholar
- Spirin, A. S. How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian ratchet model. Biochemistry48, 10688–10692 (2009). ArticleCASGoogle Scholar
- Abaeva, I. S., Pestova, T. V. & Hellen, C. U. T. Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi árva virus IRES. Nucleic Acids Res.44, 2362–2377 (2016). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hashem, Y. et al. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell153, 1108–1119 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Sweeney, T. R. et al. Functional role and ribosomal position of the unique N-terminal region of DHX29, a factor required for initiation on structured mammalian mRNAs. Nucleic Acids Res.49, 12955–12969 (2021). ArticleCASPubMedPubMed CentralGoogle Scholar
- Gao, Z. et al. Coupling between the DEAD-box RNA helicases Ded1p and eIF4A. eLife5, e16408 (2016). ArticlePubMedPubMed CentralGoogle Scholar
- Gulay, S., Gupta, N., Lorsch, J. R. & Hinnebusch, A. G. Distinct interactions of eIF4A and eIF4E with RNA helicase Ded1 stimulate translation in vivo. eLife9, e58243 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Matsuda, D. & Dreher, T. W. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA12, 1338–1349 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Rozen, F. et al. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol.10, 1134–1144 (1990). CASPubMedPubMed CentralGoogle Scholar
- Kozak, M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA87, 8301–8305 (1990). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Loughran, G. et al. Unusually efficient CUG initiation of an overlapping reading frame in POLG mRNA yields novel protein POLGARF. Proc. Natl Acad. Sci. USA117, 24936–24946 (2020). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Guenther, U.-P. et al. The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature559, 130–134 (2018). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Filipowicz, W. & Haenni, A. L. Binding of ribosomes to 5′-terminal leader sequences of eukaryotic messenger RNAs. Proc. Natl Acad. Sci. USA76, 3111–3115 (1979). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Mohammad, M. P., Smirnova, A., Gunišová, S. & Valášek, L. S. eIF4G is retained on ribosomes elongating and terminating on short upstream ORFs to control reinitiation in yeast. Nucleic Acids Res.49, 8743–8756 (2021). ArticleCASPubMedPubMed CentralGoogle Scholar
- Sonenberg, N., Shatkin, A. J., Ricciardi, R. P., Rubin, M. & Goodman, R. M. Analysis of terminal structures of RNA from potato virus X. Nucleic Acids Res.5, 2501–2512 (1978). ArticleCASPubMedPubMed CentralGoogle Scholar
- Alone, P. V., Cao, C. & Dever, T. E. Translation initiation factor 2γ mutant alters start codon selection independent of Met-tRNA binding. Mol. Cell Biol.28, 6877–6888 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Basu, I., Gorai, B., Chandran, T., Maiti, P. K. & Hussain, T. Selection of start codon during mRNA scanning in eukaryotic translation initiation. Commun. Biol.5, 587 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- Donahue, T. F., Cigan, A. M., Pabich, E. K. & Valavicius, B. C. Mutations at a Zn(II) finger motif in the yeast eIF-2β gene alter ribosomal start-site selection during the scanning process. Cell54, 621–632 (1988). ArticleCASPubMedGoogle Scholar
- Dorris, D. R., Erickson, F. L. & Hannig, E. M. Mutations in GCD11, the structural gene for eIF-2γ in yeast, alter translational regulation of GCN4 and the selection of the start site for protein synthesis. EMBO J.14, 2239–2249 (1995). ArticleCASPubMedPubMed CentralGoogle Scholar
- Das, S., Ghosh, R. & Maitra, U. Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J. Biol. Chem.276, 6720–6726 (2001). ArticleCASPubMedGoogle Scholar
- Paulin, F. E. M., Campbell, L. E., O’Brien, K., Loughlin, J. & Proud, C. G. Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase-activator protein. Curr. Biol.11, 55–59 (2001). ArticleCASPubMedGoogle Scholar
- Algire, M. A., Maag, D. & Lorsch, J. R. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell20, 251–262 (2005). This study presents the first evidence that Pi release and not GTP hydrolysis marks the end of scanning.ArticleCASPubMedGoogle Scholar
- Simonetti, A. et al. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol. Cell63, 206–217 (2016). ArticleCASPubMedGoogle Scholar
- Yu, Y. et al. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res.37, 5167–5182 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kozak, M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature308, 241–246 (1984). ArticleADSCASPubMedGoogle Scholar
- Acker, M. G., Shin, B.-S., Dever, T. E. & Lorsch, J. R. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem.281, 8469–8475 (2006). ArticleCASPubMedGoogle Scholar
- Brown, Z. P. et al. Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES. EMBO J.41, e110581 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- Fringer, J. M., Acker, M. G., Fekete, C. A., Lorsch, J. R. & Dever, T. E. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell Biol.27, 2384–2397 (2007). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kazan, R. et al. Role of aIF5B in archaeal translation initiation. Nucleic Acids Res.50, 6532–6548 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lee, J. H. et al. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl Acad. Sci. USA99, 16689–16694 (2002). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Acker, M. G. et al. Kinetic analysis of late steps of eukaryotic translation initiation. J. Mol. Biol.385, 491–506 (2009). ArticleCASPubMedGoogle Scholar
- Fernández, I. S. et al. Molecular architecture of a eukaryotic translational initiation complex. Science342, 1240585 (2013). ArticlePubMedGoogle Scholar
- Shin, B.-S. et al. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell111, 1015–1025 (2002). ArticleCASPubMedGoogle Scholar
- Unbehaun, A., Borukhov, S. I., Hellen, C. U. T. & Pestova, T. V. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev.18, 3078–3093 (2004). ArticleCASPubMedPubMed CentralGoogle Scholar
- Wagner, S. et al. Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. Mol. Cell79, 546–560.e7 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lin, Y. et al. eIF3 associates with 80S ribosomes to promote translation elongation, mitochondrial homeostasis, and muscle health. Mol. Cell79, 575–587.e7 (2020). ArticleCASPubMedGoogle Scholar
- Mohammad, M. P., Munzarová Pondelícková, V., Zeman, J., Gunišová, S. & Valášek, L. S. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res.45, 2658–2674 (2017). CASPubMedPubMed CentralGoogle Scholar
- Munzarová, V. et al. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet.7, e1002137 (2011). ArticlePubMedPubMed CentralGoogle Scholar
- Jang, S. K. et al. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol.62, 2636–2643 (1988). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pelletier, J. & Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature334, 320–325 (1988). ArticleADSCASPubMedGoogle Scholar
- Lee, K.-M., Chen, C.-J. & Shih, S.-R. Regulation mechanisms of viral IRES-driven translation. Trends Microbiol.25, 546–561 (2017). ArticleCASPubMedGoogle Scholar
- Filbin, M. E., Vollmar, B. S., Shi, D., Gonen, T. & Kieft, J. S. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat. Struct. Mol. Biol.20, 150–158 (2013). ArticleCASPubMedGoogle Scholar
- Hashem, Y. et al. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature503, 539–543 (2013). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Murray, J. et al. Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife5, e13567 (2016). ArticlePubMedPubMed CentralGoogle Scholar
- Neupane, R., Pisareva, V. P., Rodriguez, C. F., Pisarev, A. V. & Fernández, I. S. A complex IRES at the 5′-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate. eLife9, e54575 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Quade, N., Boehringer, D., Leibundgut, M., van den Heuvel, J. & Ban, N. Cryo-EM structure of hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat. Commun.6, 7646 (2015). ArticleADSPubMedGoogle Scholar
- Schüler, M. et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol.13, 1092–1096 (2006). ArticlePubMedGoogle Scholar
- Yamamoto, H. et al. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol.21, 721–727 (2014). ArticleCASPubMedGoogle Scholar
- Hellen, C. U. & Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev.15, 1593–1612 (2001). ArticleCASPubMedGoogle Scholar
- Macejak, D. G. & Sarnow, P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature353, 90–94 (1991). ArticleADSCASPubMedGoogle Scholar
- Stoneley, M. & Willis, A. E. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene23, 3200–3207 (2004). ArticleCASPubMedGoogle Scholar
- Ivanov, I. P. et al. Evolutionarily conserved inhibitory uORFs sensitize Hox mRNA translation to start codon selection stringency. Proc. Natl Acad. Sci. USA119, e2117226119 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature495, 384–388 (2013). ArticleADSCASPubMedGoogle Scholar
- Kleaveland, B., Shi, C. Y., Stefano, J. & Bartel, D. P. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell174, 350–362.e17 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature495, 333–338 (2013). ArticleADSCASPubMedGoogle Scholar
- Di Timoteo, G. et al. Modulation of circRNA Metabolism by m 6 A modification. Cell Rep.31, 107641 (2020). ArticlePubMedGoogle Scholar
- Perry, R. P., Kelley, D. E., Friderici, K. & Rottman, F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell4, 387–394 (1975). ArticleCASPubMedGoogle Scholar
- Viegas, I. J. et al. N 6 -Methyladenosine in poly(A) tails stabilize VSG transcripts. Naturehttps://doi.org/10.1038/s41586-022-04544-0 (2022). ArticlePubMedPubMed CentralGoogle Scholar
- Wang, X. et al. N 6 -Methyladenosine-dependent regulation of messenger RNA stability. Nature505, 117–120 (2014). ArticleADSPubMedGoogle Scholar
- Su, R. et al. METTL16 exerts an m 6 A-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol.24, 205–216 (2022). ArticleMathSciNetCASPubMedPubMed CentralGoogle Scholar
- Wang, X. et al. Structural basis of N 6 -adenosine methylation by the METTL3–METTL14 complex. Nature534, 575–578 (2016). ArticleADSCASPubMedGoogle Scholar
- Li, A. et al. Cytoplasmic m 6 A reader YTHDF3 promotes mRNA translation. Cell Res.27, 444–447 (2017). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Takahashi, H., Kato, S., Murata, M. & Carninci, P. CAGE—cap analysis gene expression: a protocol for the detection of promoter and transcriptional networks. Methods Mol. Biol.786, 181–200 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Jia, L. & Qian, S.-B. A versatile eIF3d in translational control of stress adaptation. Mol. Cell81, 10–12 (2021). ArticleCASPubMedGoogle Scholar
- de la Parra, C. et al. A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun.9, 3068 (2018). ArticleADSPubMedPubMed CentralGoogle Scholar
- Volta, V. et al. A DAP5/eIF3d alternate mRNA translation mechanism promotes differentiation and immune suppression by human regulatory T cells. Nat. Commun.12, 6979 (2021). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Haizel, S. A., Bhardwaj, U., Gonzalez, R. L., Mitra, S. & Goss, D. J. 5′-UTR recruitment of the translation initiation factor eIF4GI or DAP5 drives cap-independent translation of a subset of human mRNAs. J. Biol. Chem.295, 11693–11706 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Costa-Mattioli, M. & Walter, P. The integrated stress response: from mechanism to disease. Science368, eaat5314 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pakos-Zebrucka, K. et al. The integrated stress response. EMBO Rep.17, 1374–1395 (2016). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hinnebusch, A. G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol.59, 407–450 (2005). ArticleCASPubMedGoogle Scholar
- Hinnebusch, A. G., Ivanov, I. P. & Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science352, 1413–1416 (2016). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Bohlen, J. et al. DENR promotes translation reinitiation via ribosome recycling to drive expression of oncogenes including ATF4. Nat. Commun.11, 4676 (2020). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Skabkin, M. A. et al. Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev.24, 1787–1801 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lomakin, I. B., Dmitriev, S. E. & Steitz, T. A. Crystal structure of the DENR–MCT-1 complex revealed zinc-binding site essential for heterodimer formation. Proc. Natl Acad. Sci. USA116, 528–533 (2019). ArticleADSCASPubMedGoogle Scholar
- Weisser, M. et al. Structural and functional insights into human re-initiation complexes. Mol. Cell67, 447–456.e7 (2017). ArticleCASPubMedGoogle Scholar
- Green, K. M., Miller, S. L., Malik, I. & Todd, P. K. Non-canonical initiation factors modulate repeat-associated non-AUG translation. Hum. Mol. Genet.https://doi.org/10.1093/hmg/ddac021 (2022). ArticlePubMedPubMed CentralGoogle Scholar
- Komar, A. A. & Merrick, W. C. A retrospective on eIF2A- and not the α subunit of eIF2. Int. J. Mol. Sci.21, 2054 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Tahmasebi, S., Khoutorsky, A., Mathews, M. B. & Sonenberg, N. Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol.19, 791–807 (2018). ArticleCASPubMedGoogle Scholar
- Robichaud, N., Sonenberg, N., Ruggero, D. & Schneider, R. J. Translational control in cancer. Cold Spring Harb. Perspect. Biol.11, a032896 (2019). ArticleCASPubMedPubMed CentralGoogle Scholar
- Alvarez, E., Menéndez-Arias, L. & Carrasco, L. The eukaryotic translation initiation factor 4GI is cleaved by different retroviral proteases. J. Virol.77, 12392–12400 (2003). ArticleCASPubMedPubMed CentralGoogle Scholar
- Etchison, D., Milburn, S. C., Edery, I., Sonenberg, N. & Hershey, J. W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem.257, 14806–14810 (1982). ArticleCASPubMedGoogle Scholar
- Ventoso, I., Blanco, R., Perales, C. & Carrasco, L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl Acad. Sci. USA98, 12966–12971 (2001). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Kamitani, W., Huang, C., Narayanan, K., Lokugamage, K. G. & Makino, S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol.16, 1134–1140 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kamitani, W. et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl Acad. Sci. USA103, 12885–12890 (2006). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Yuan, S. et al. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol. Cell80, 1055–1066.e6 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lapointe, C. P. et al. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc. Natl Acad. Sci. USA118, e2017715118 (2021). ArticleCASPubMedPubMed CentralGoogle Scholar
- Mendez, A. S. et al. The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep.37, 109841 (2021). ArticleCASPubMedPubMed CentralGoogle Scholar
- Rao, S. et al. Genes with 5′ terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 Nsp1 protein. RNA27, 1025–1045 (2021). ArticleCASPubMedPubMed CentralGoogle Scholar
- Sosnowski, P., Tidu, A., Eriani, G., Westhof, E. & Martin, F. Correlated sequence signatures are present within the genomic 5′ UTR RNA and NSP1 protein in coronaviruses. RNAhttps://doi.org/10.1261/rna.078972.121 (2022). ArticlePubMedPubMed CentralGoogle Scholar
- Vora, S. M. et al. Targeting stem-loop 1 of the SARS-CoV-2 5′ UTR to suppress viral translation and Nsp1 evasion. Proc. Natl Acad. Sci. USA119, e2117198119 (2022). ArticleCASPubMedPubMed CentralGoogle Scholar
- Tanaka, T., Kamitani, W., DeDiego, M. L., Enjuanes, L. & Matsuura, Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol.86, 11128–11137 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Cleary, J. D., Pattamatta, A. & Ranum, L. P. W. Repeat-associated non-ATG (RAN) translation. J. Biol. Chem.293, 16127–16141 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Storkebaum, E., Rosenblum, K. & Sonenberg, N. Messenger RNA translation defects in neurodegenerative diseases. N. Engl. J. Med.388, 1015–1030 (2023). ArticleCASPubMedGoogle Scholar
- Ash, P. E. A. et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron77, 639–646 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Green, K. M. et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun.8, 2005 (2017). ArticleADSPubMedPubMed CentralGoogle Scholar
- Mori, K. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science339, 1335–1338 (2013). ArticleADSCASPubMedGoogle Scholar
- Tabet, R. et al. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat. Commun.9, 152 (2018). ArticleADSPubMedPubMed CentralGoogle Scholar
- Zu, T. et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl Acad. Sci. USA108, 260–265 (2011). ArticleADSCASPubMedGoogle Scholar
- Afonja, O., Juste, D., Das, S., Matsuhashi, S. & Samuels, H. H. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene23, 8135–8145 (2004). ArticleCASPubMedGoogle Scholar
- Chen, Y. et al. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J. Pathol.200, 640–646 (2003). ArticleCASPubMedGoogle Scholar
- Zhang, H. et al. Involvement of programmed cell death 4 in transforming growth factor-β1-induced apoptosis in human hepatocellular carcinoma. Oncogene25, 6101–6112 (2006). ArticleCASPubMedGoogle Scholar
- Biyanee, A., Ohnheiser, J., Singh, P. & Klempnauer, K.-H. A novel mechanism for the control of translation of specific mRNAs by tumor suppressor protein Pdcd4: inhibition of translation elongation. Oncogene34, 1384–1392 (2015). ArticleCASPubMedGoogle Scholar
- Singh, P., Wedeken, L., Waters, L. C., Carr, M. D. & Klempnauer, K.-H. Pdcd4 directly binds the coding region of c-myb mRNA and suppresses its translation. Oncogene30, 4864–4873 (2011). ArticleCASPubMedGoogle Scholar
- Safaee, N. et al. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol. Cell48, 375–386 (2012). ArticleCASPubMedGoogle Scholar
- Grüner, S. et al. The structures of eIF4E–eIF4G complexes reveal an extended interface to regulate translation initiation. Mol. Cell64, 467–479 (2016). ArticlePubMedGoogle Scholar
- Schütz, P. et al. Crystal structure of the yeast eIF4A–eIF4G complex: an RNA-helicase controlled by protein–protein interactions. Proc. Natl Acad. Sci. USA105, 9564–9569 (2008). ArticleADSPubMedPubMed CentralGoogle Scholar
- Yang, M., Lu, Y., Piao, W. & Jin, H. The translational regulation in mTOR pathway. Biomolecules12, 802 (2022). ArticlePubMedPubMed CentralGoogle Scholar
- Hernández, G. The versatile relationships between eIF4E and eIF4E-interacting proteins. Trends Genet.38, 801–804 (2022). ArticlePubMedGoogle Scholar
- Berman, A. J. et al. Controversies around the function of LARP1. RNA Biol.18, 207–217 (2021). ArticleCASPubMedGoogle Scholar
- Meyuhas, O. & Kahan, T. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta1849, 801–811 (2015). ArticleCASPubMedGoogle Scholar
- Christie, M. & Igreja, C. eIF4E-homologous protein (4EHP): a multifarious cap-binding protein. FEBS J.https://doi.org/10.1111/febs.16275 (2021). ArticlePubMedGoogle Scholar
- Adomavicius, T. et al. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun.10, 2136 (2019). ArticleADSPubMedPubMed CentralGoogle Scholar
- Gordiyenko, Y., Llácer, J. L. & Ramakrishnan, V. Structural basis for the inhibition of translation through eIF2α phosphorylation. Nat. Commun.10, 2640 (2019). ArticleADSPubMedPubMed CentralGoogle Scholar
- Kashiwagi, K. et al. Structural basis for eIF2B inhibition in integrated stress response. Science364, 495–499 (2019). ArticleADSCASPubMedGoogle Scholar
- Kenner, L. R. et al. eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response. Science364, 491–495 (2019). ArticleADSCASPubMedPubMed CentralGoogle Scholar
- Krishnamoorthy, T., Pavitt, G. D., Zhang, F., Dever, T. E. & Hinnebusch, A. G. Tight binding of the phosphorylated α subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell Biol.21, 5018–5030 (2001). ArticleCASPubMedPubMed CentralGoogle Scholar
- Tsai, J. C. et al. Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science359, eaaq0939 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Zyryanova, A. F. et al. Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B. Science359, 1533–1536 (2018). ArticleADSCASPubMedPubMed CentralGoogle Scholar
Acknowledgements
The authors thank T. Dever, A. Hinnebusch, J. Lorsch, C. S. Fraser, N. Sonenberg, J. Pelletier and W. Filipowicz for feedback on the manuscript. J.B.Q. was supported by a Federation of European Biochemical Societies long-term fellowship; I.D.-L. was supported by an EMBO Postdoctoral Fellowship; and V.R. was supported by the UK Medical Research Council (MC_U105184332), a Wellcome Trust Investigator award (WT096570) and the Louis-Jeantet Foundation.










